Tutorial¶
- Download the sample input file
$ curl -O http://paoyang.ipmb.sinica.edu.tw/public_data/methgo_demo.tar.gz $ tar xvfz methgo_demo.tar.gz $ cd methgo_demo/data
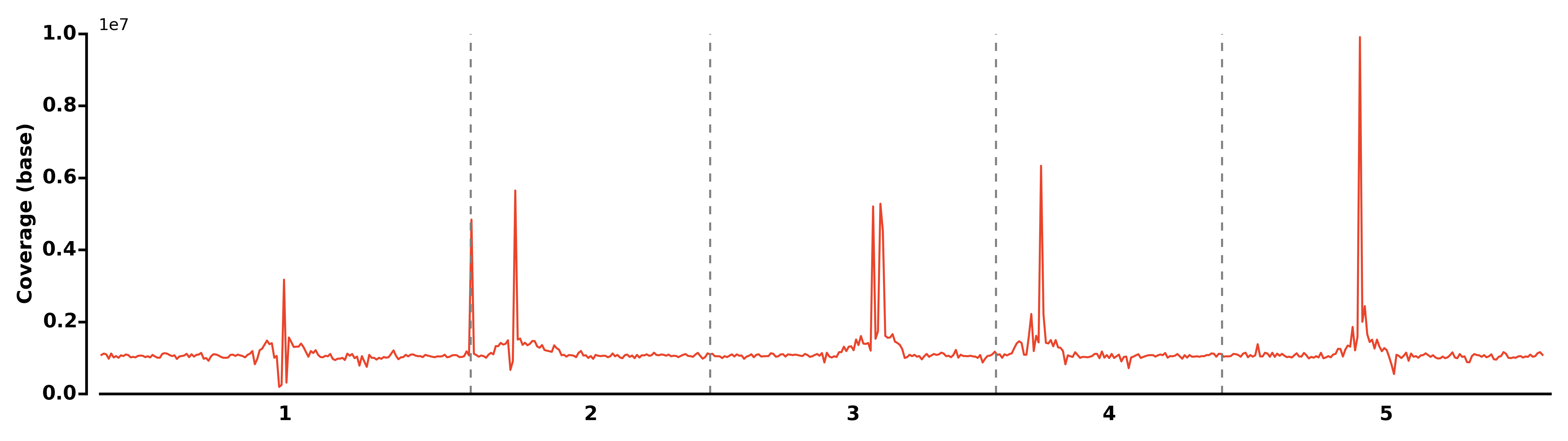
- Run COV module:
$ methgo cov genome.fa demo.CGmap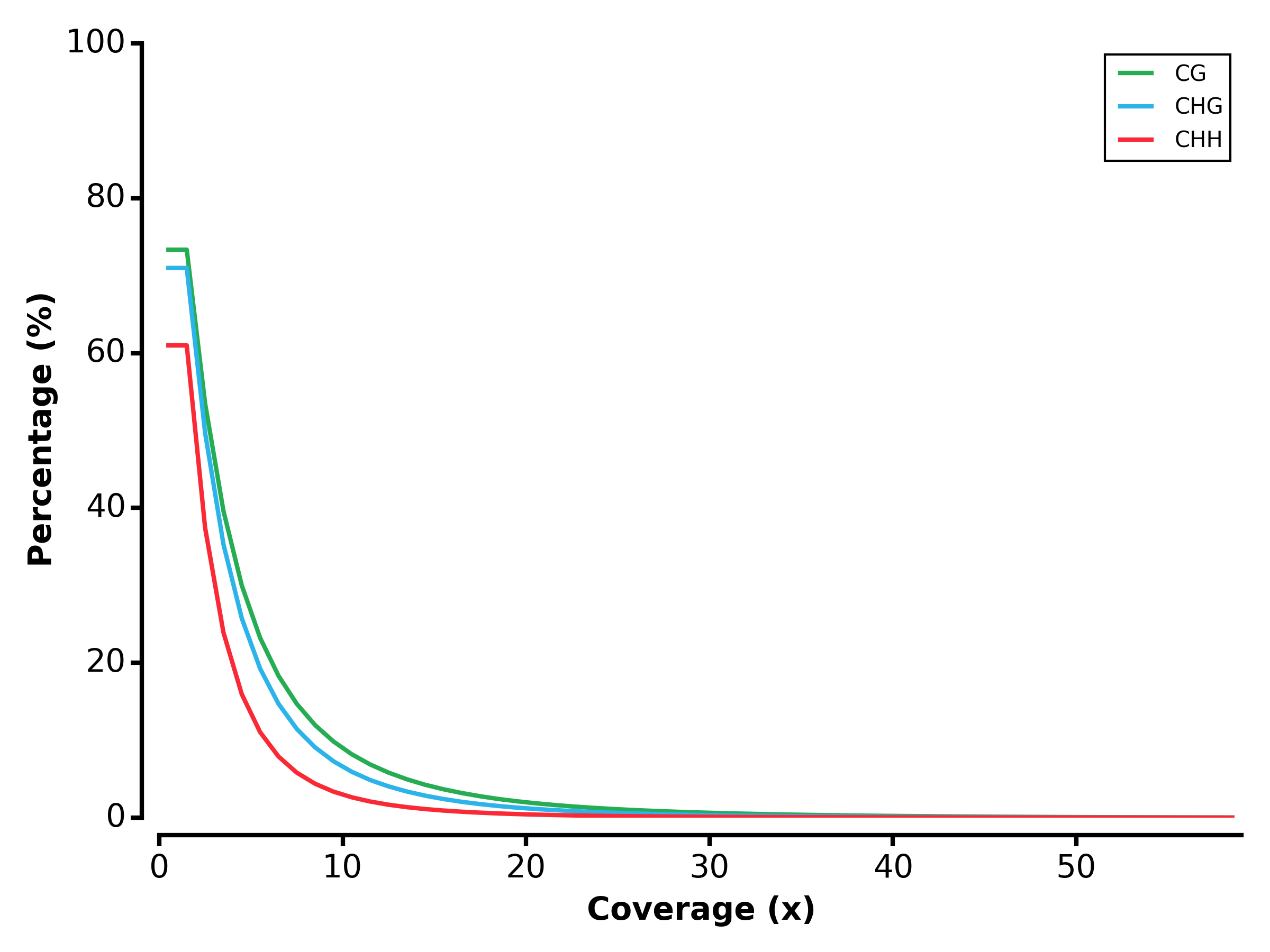
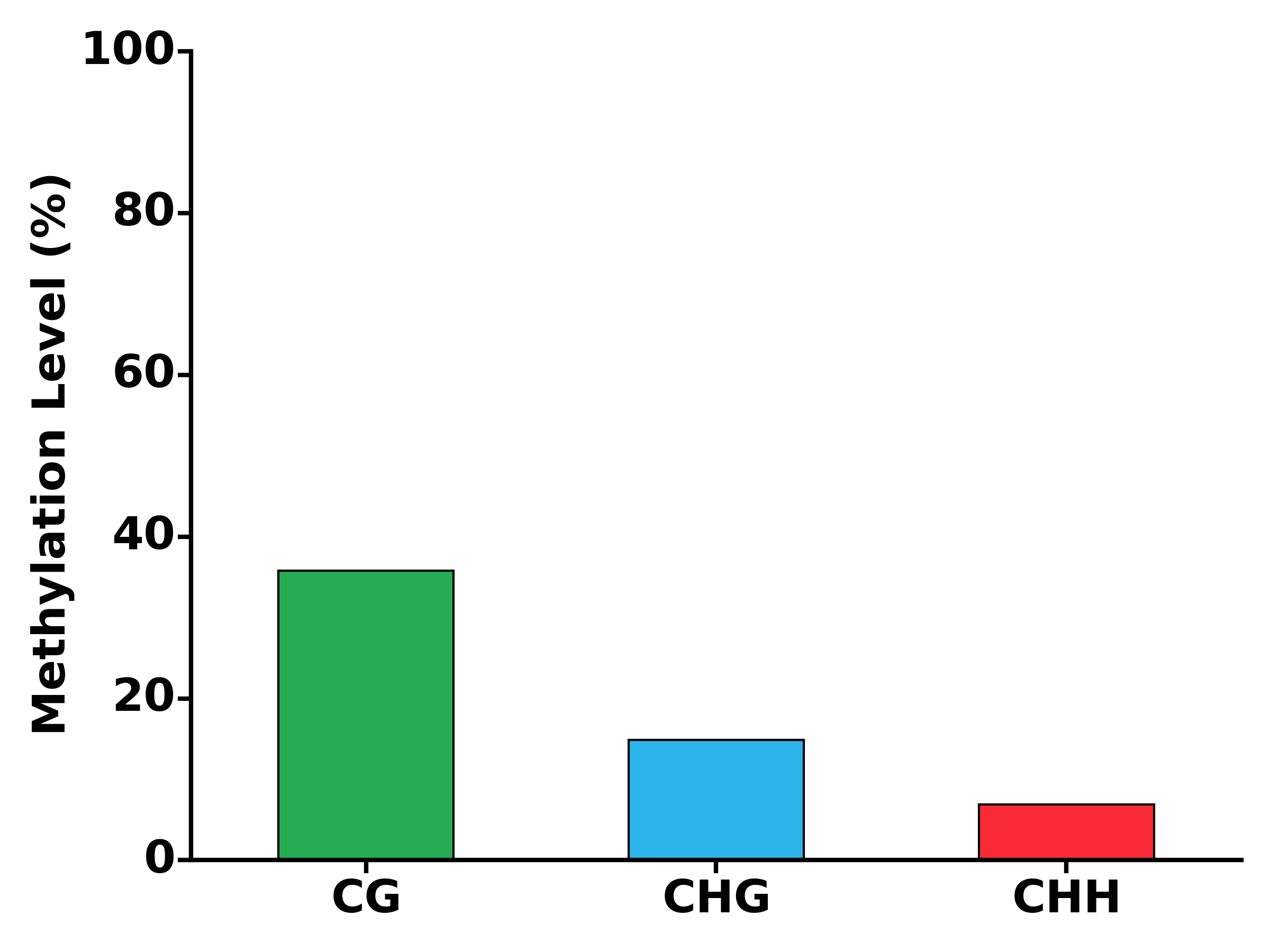
- Run MET module:
$ methgo met genes.gtf genome.fa demo.CGmap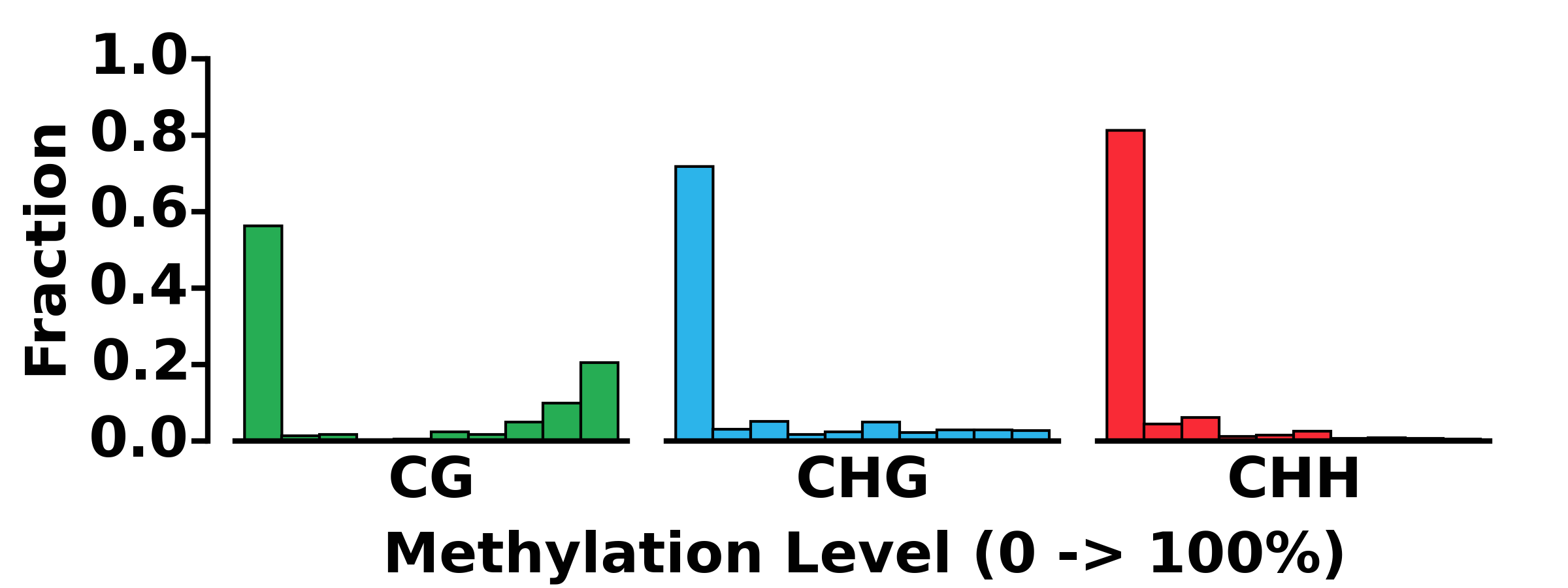
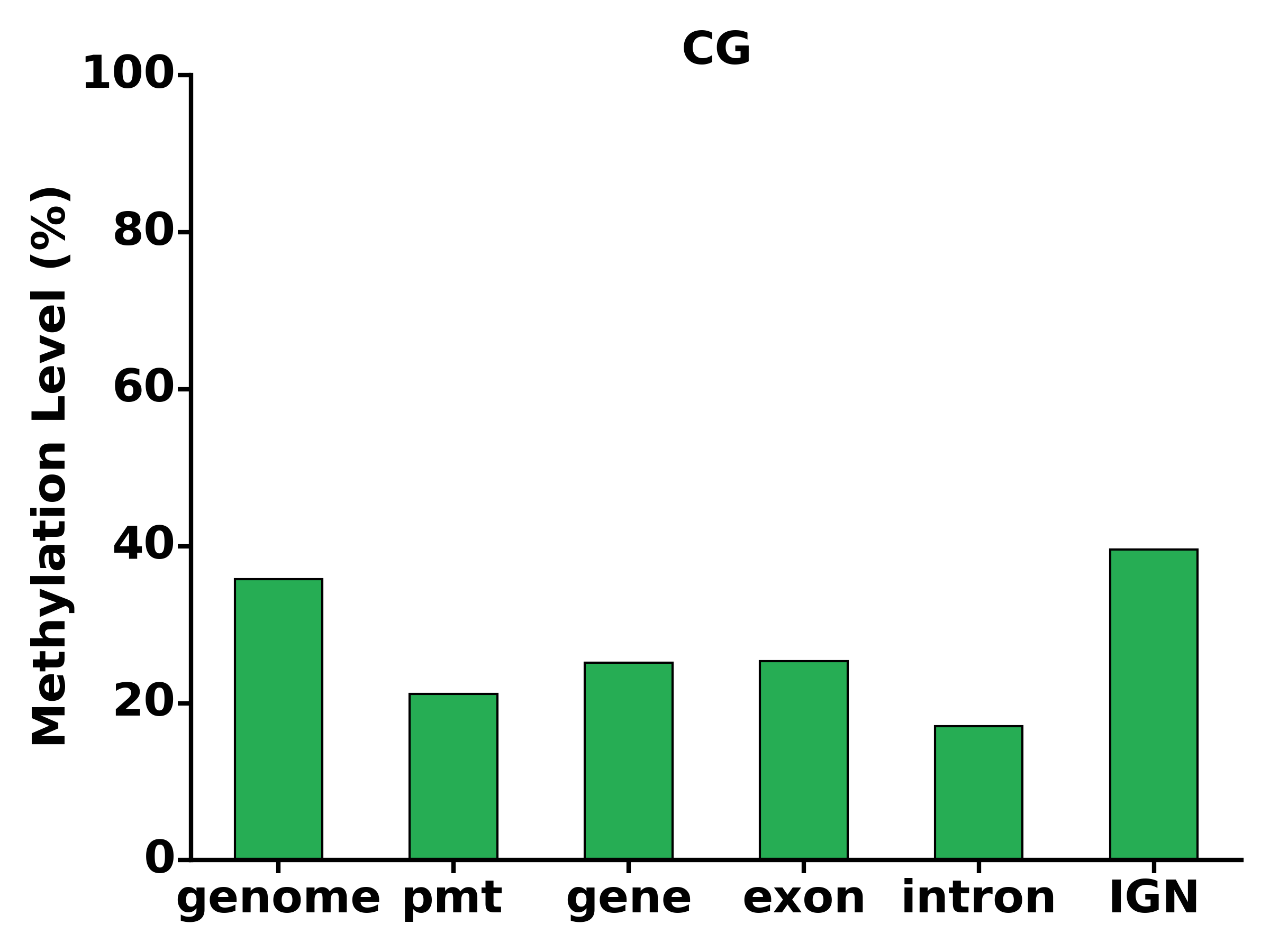
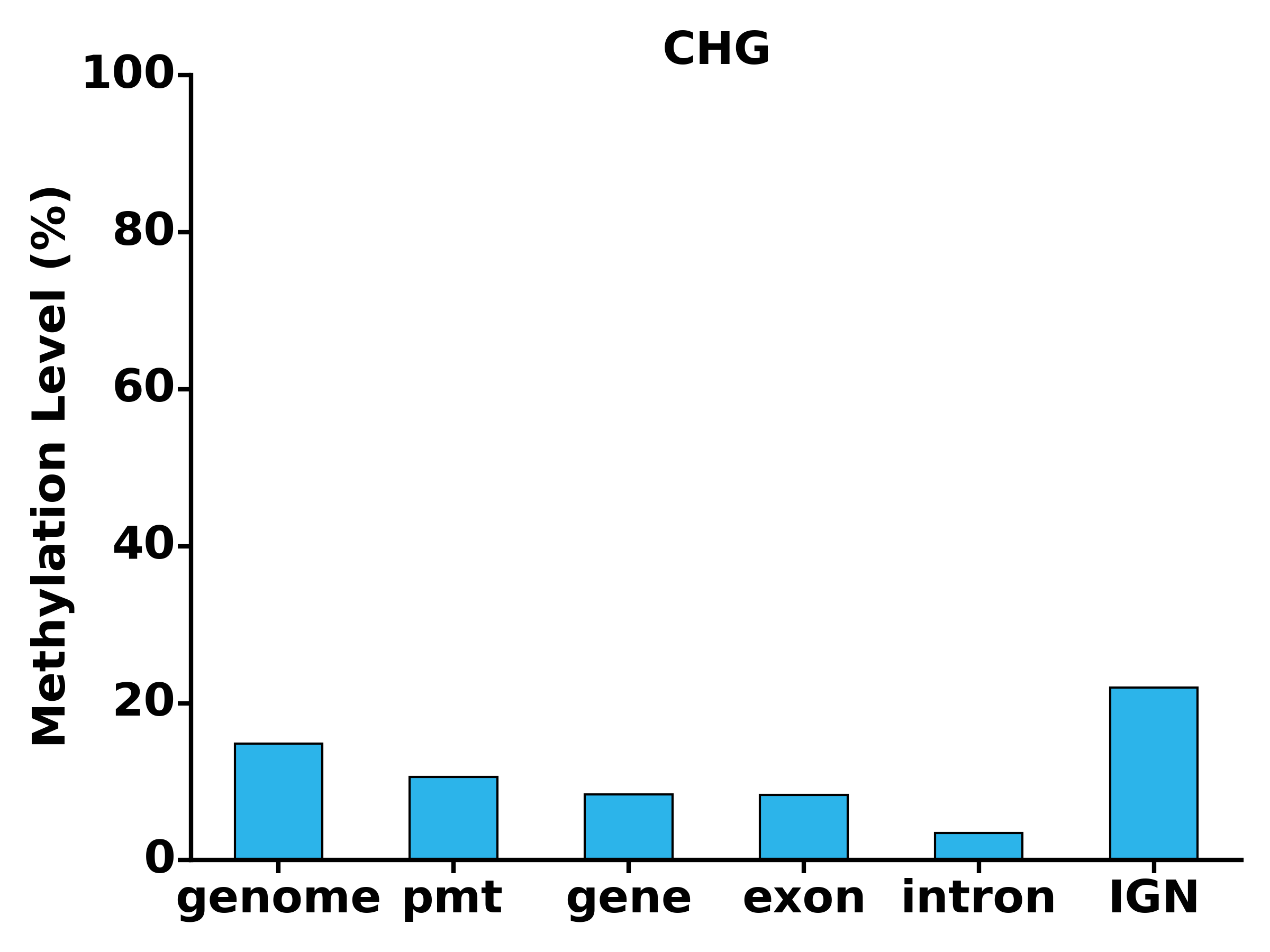
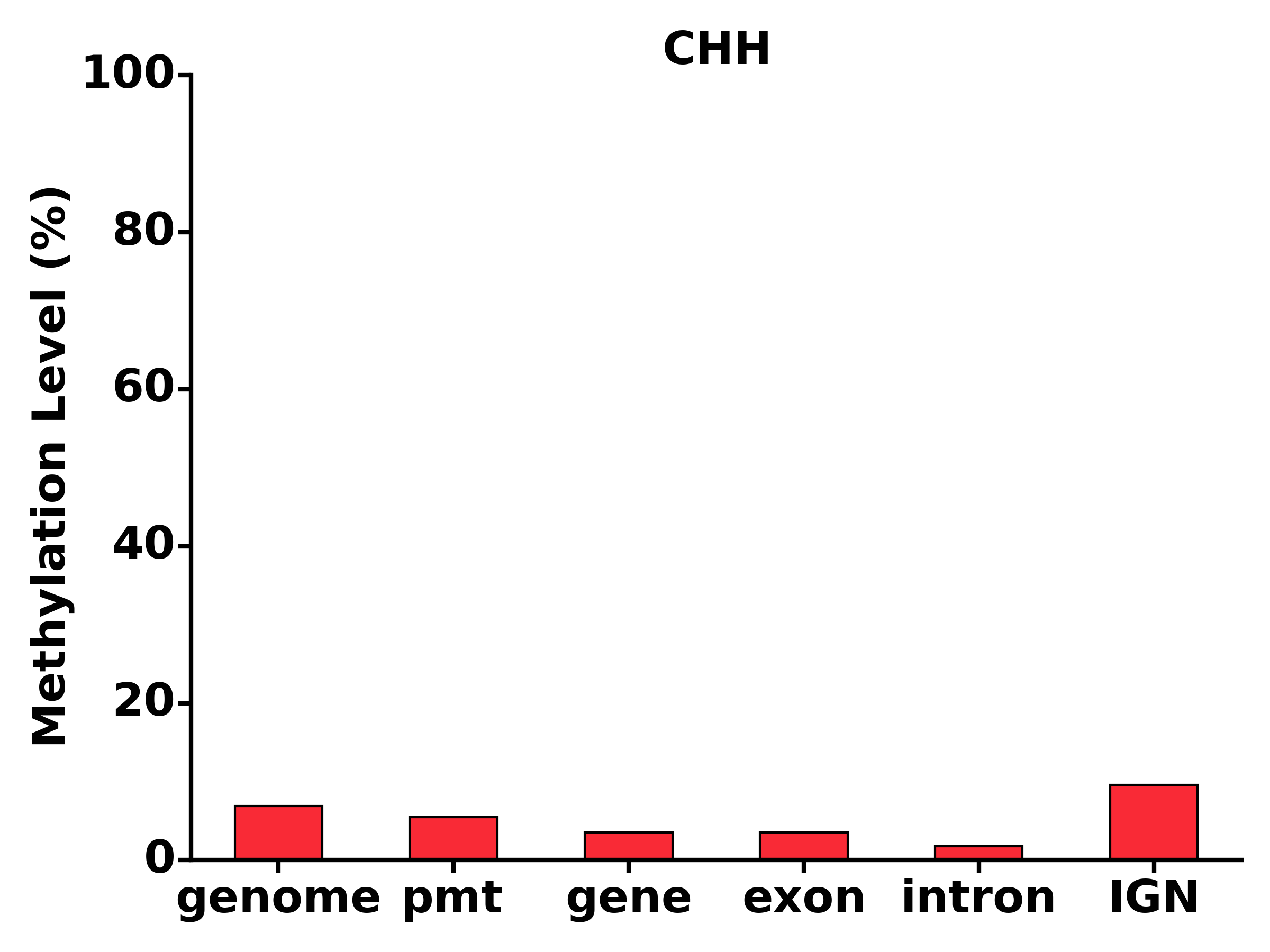
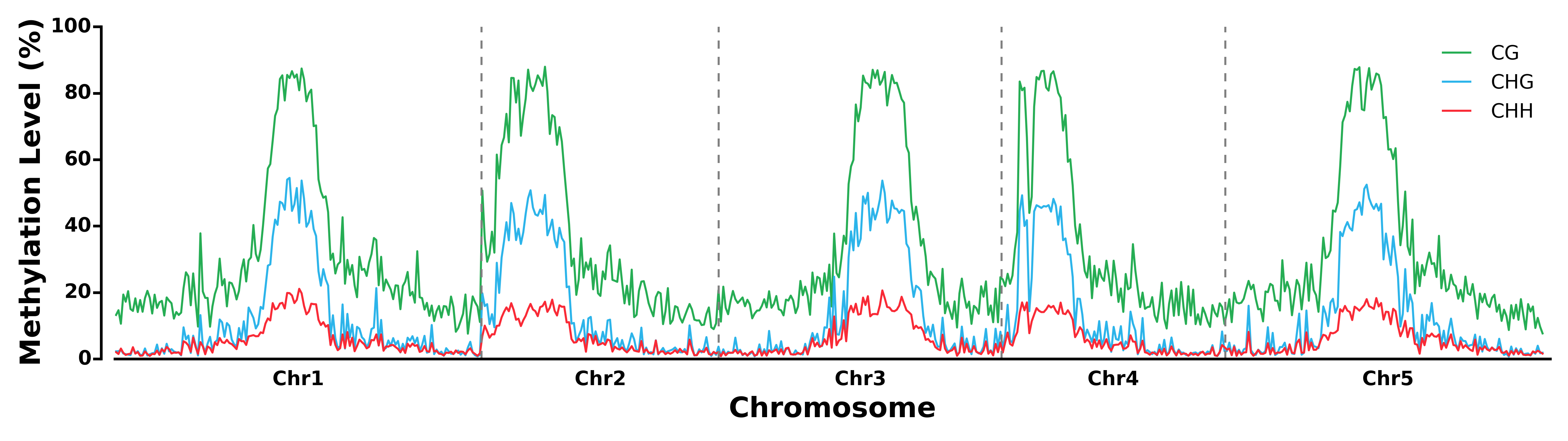
- Run TXN module:
$ methgo txn -t methgo/scripts/txn/tair10_txn -l ATHB1_binding_site_motif,CCA1_binding_site_motif -c demo.CGmap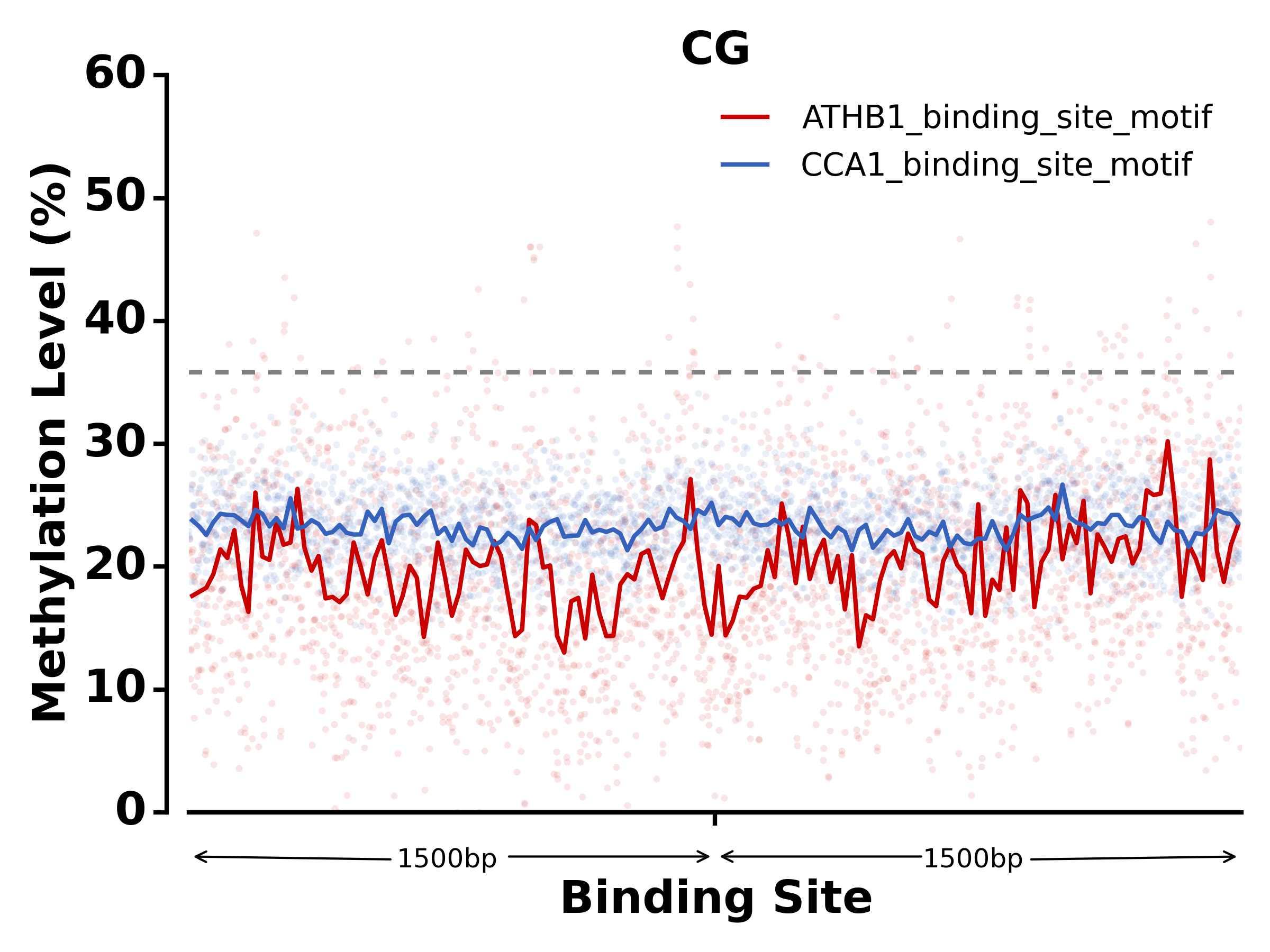
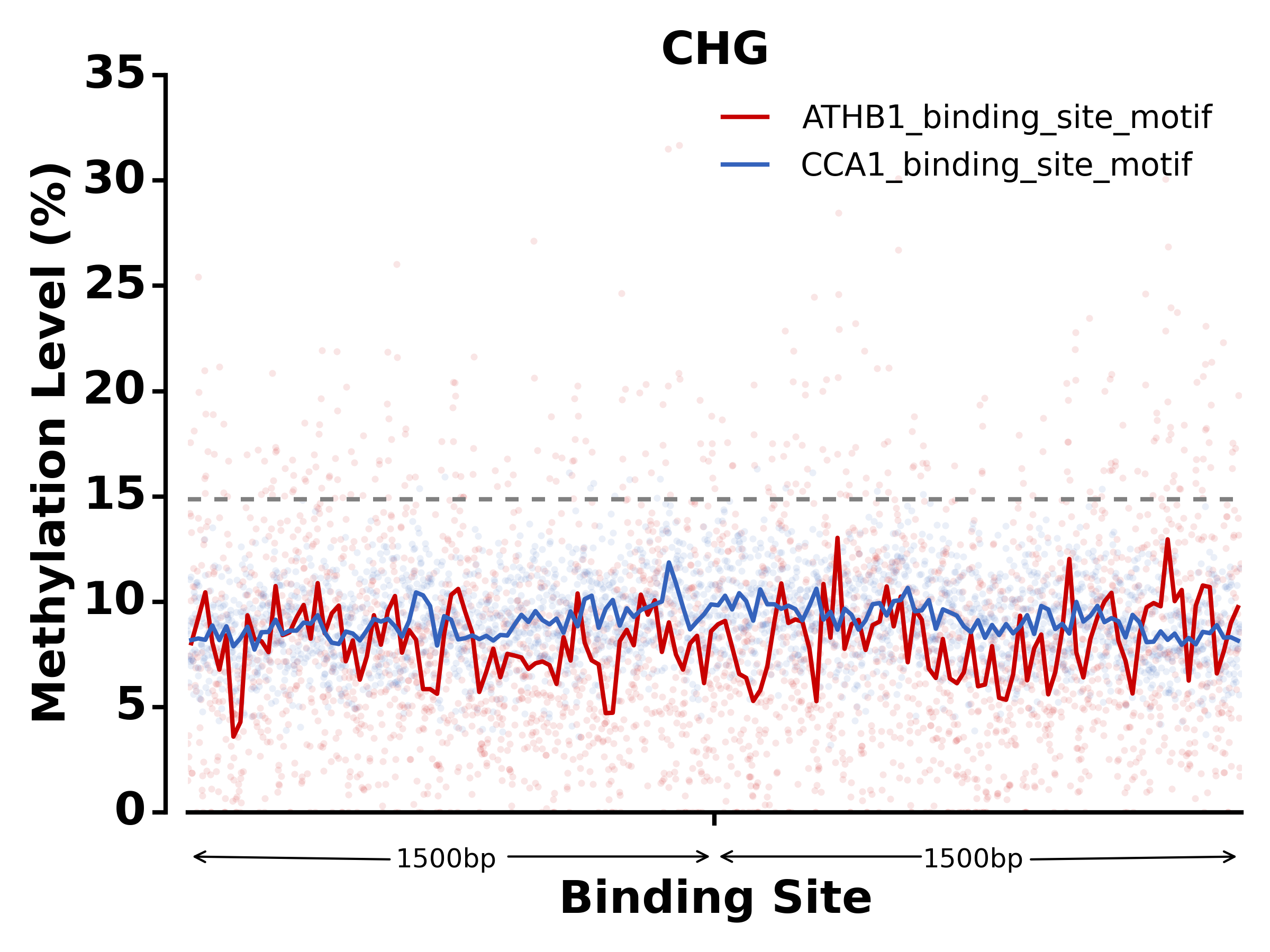
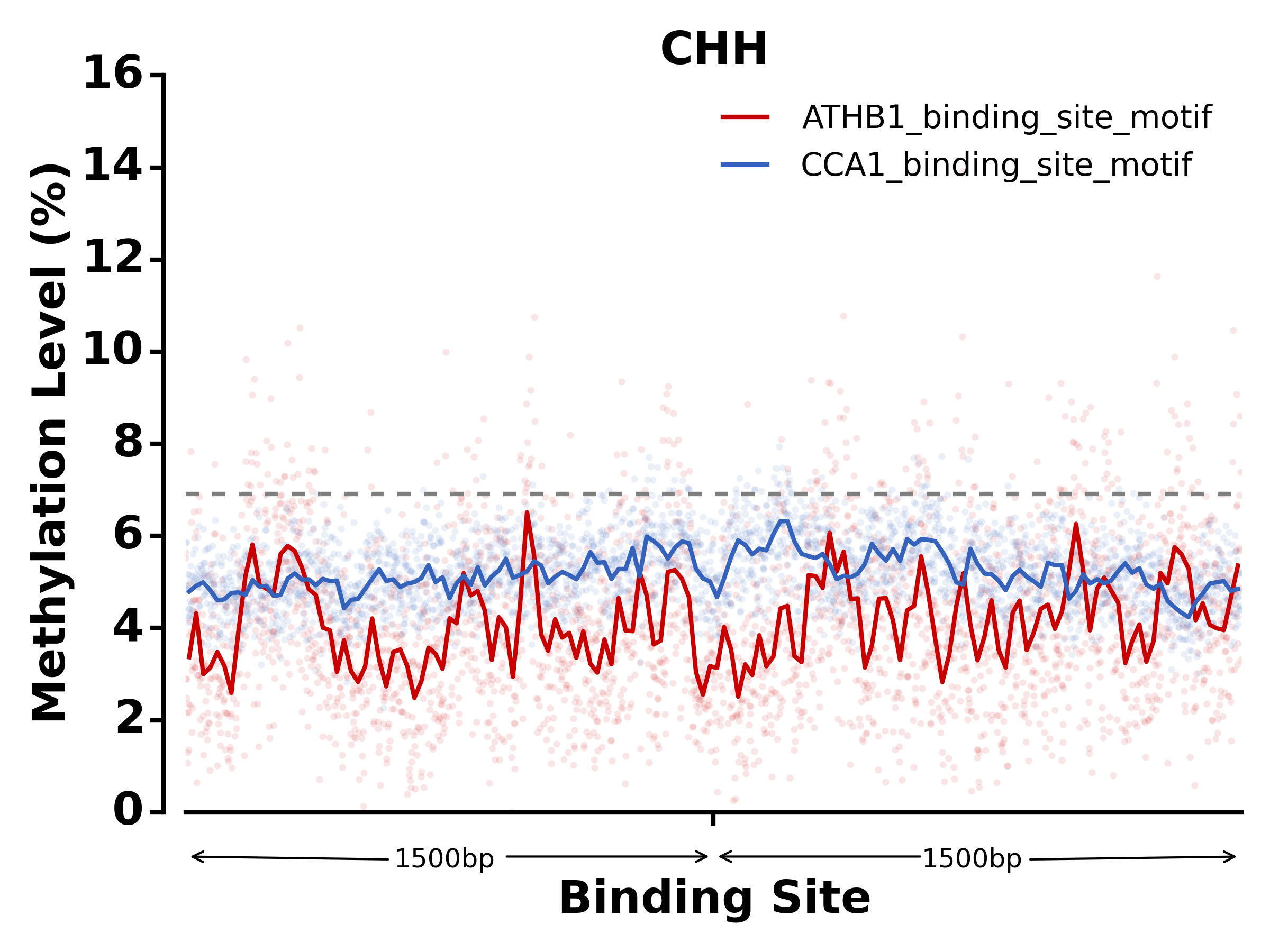
- Run SNP module:
$ methgo snp -g genome.fa demo.sorted.bam
- Run CNV module:
$ methgo cnv genome.fa.fai demo.sorted.bam